Example results
This page contains some example results from a previously prepared Companion annotation run. The tabs below display live data and behave exactly like an actual job result page (in fact, they are the result of an actual job that was run over 2 years ago!). Click the tabs to see what data they contain.
| Reference | Query | |
|---|---|---|
| Total number of regions/sequences | 9 | 8 |
| Number of annotated regions/sequences | 9 | |
| Number of genes | 6459 | 5930 |
| Gene density (genes/megabase) | – | 399.02 |
| Number of coding genes | 6212 | 5833 |
| Number of pseudogenes | – | 193 |
| Number of genes with function | – | 5734 |
| Number of pseudogenes with function | – | 190 |
| Number of non-coding genes | 247 | 97 |
| Number of genes with multiple CDSs | – | 448 |
| Overall GC% | – | 33.25 |
| Coding GC% | – | 34.89 |
| Format | MD5 | Size | |
|---|---|---|---|
| Pseudochromosome level genomic sequence | FASTA | 4.06 MB | |
| Pseudochromosome level gene annotations | GFF3 | 4.88 MB | |
| Pseudochromosome layout | AGP | 362 Bytes | |
| Scaffold level genomic sequence | FASTA | 4.06 MB | |
| Scaffold level gene annotations | GFF3 | 4.91 MB | |
| Scaffold layout | AGP | 418 Bytes | |
| Pseudochromosome level sequence and annotation | EMBL | 7.86 MB | |
| Gene Ontology function assignments | GAF1 | 14.4 MB | |
| Protein sequences | FASTA | 3.45 MB |

These clusters have been created using OrthoFinder v2.5.4 based on Diamond hits (e-value cutoff: 10-3) between the translated protein sequences of the target and reference genomes' protein-coding genes.
Cluster assignments as OrthoFinder Orthogroups output file
Click and drag in the diagram below to pan around. Use the mouse wheel to zoom in and out. The newly annotated genome in this job is highlighted: CDUB.

These clusters have been created using FastTree based on a multiple sequence alignment created by MAFFT (--auto --anysymbol --retree 1 --parttree) of the concatenated DNA sequences of up to 50 single-copy core genes from all organisms appearing as taxa in the tree above.
Tree drawn by Phylocanvas.
Multiple sequence alignment
for this tree (FASTA)
Core genes
used to build this tree
Each circle below represents a single target-reference pseudochromosome alignment. Click on the thumbnail to zoom in. Download all 8 images (ZIP)
Standard output:
N E X T F L O W ~ version 21.10.6
Launching `/home/companion/annot-nf/annot.nf` [stupefied_heyrovsky] - revision: d107eedd43
WARN: It appears you have never run this project before -- Option `-resume` is ignored
C O M P A N I O N ~ v2.0.10
query : /www/companion/annot-web/public/system/dragonfly/production/2023/07/19/4qok0frcmh_FungiDB_60_CdubliniensisCD36_Genome.fasta
reference : Candida_albicans_SC5314
reference directory : /data/references/ref_fungidb/Ref_Candida
output directory : /www/companion/annot-web/public/jobs/258ea8f6468bb5809c5056ac
WARN: The operator `first` is useless when applied to a value channel which returns a single value by definition -- check channel `ncrna_cmindex`
WARN: Access to undefined parameter `TRANSCRIPT_FILE` -- Initialise it to a default value eg. `params.TRANSCRIPT_FILE = some_value`
WARN: The access of `config` object is deprecated
[bf/246116] Submitted process > press_ncRNA_cms
WARN: The `into` operator should be used to connect two or more target channels -- consider to replace it with `.set { integrated_gff3_processed }`
WARN: The operator `first` is useless when applied to a value channel which returns a single value by definition -- check channel `pseudochr_last_index`
[fb/90d65c] Submitted process > truncate_input_headers
[c7/b9ac94] Submitted process > exonerate_empty_hints
[f0/605100] Submitted process > transcript_empty_hints
[c7/974c5a] Submitted process > ratt_make_ref_embl
[08/b5f055] Submitted process > make_empty_snap
[b2/1c8ea0] Submitted process > pseudogene_indexing
[e4/7efe8b] Submitted process > make_ref_input_for_orthomcl
[82/6d6b7a] Submitted process > merge_hints
[b2/63293d] Submitted process > sanitize_input
[ad/86a2cc] Submitted process > contiguate_pseudochromosomes
[95/ee468c] Submitted process > nucmer_for_circos (5)
[88/9e8c3d] Submitted process > nucmer_for_circos (9)
[97/03073c] Submitted process > predict_tRNA
[79/61ae80] Submitted process > nucmer_for_circos (3)
[e2/add56b] Submitted process > nucmer_for_circos (1)
[47/2d90cb] Submitted process > nucmer_for_circos (8)
[00/e42b4a] Submitted process > make_distribution_seqs
[ae/92151d] Submitted process > pseudogene_last (3)
[79/61c9c0] Submitted process > nucmer_for_circos (6)
[b3/5214cb] Submitted process > nucmer_for_circos (2)
[92/c1a594] Submitted process > nucmer_for_circos (7)
[9d/aed2e5] Submitted process > nucmer_for_circos (4)
[97/e921e2] Submitted process > run_ratt
[4d/02f6fb] Submitted process > run_braker_pseudo
[63/6c78ae] Submitted process > predict_ncRNA (1)
[94/f2a31f] Submitted process > pseudogene_last (1)
[11/7a5ee9] Submitted process > pseudogene_last (2)
[44/bb17a7] Submitted process > predict_ncRNA (2)
WARN: Access to undefined parameter `print_paths` -- Initialise it to a default value eg. `params.print_paths = some_value`
[3c/bd678a] Submitted process > ratt_to_gff3
[d7/5607cb] Submitted process > merge_ncrnas (1)
[20/6f1aff] Submitted process > parse_braker_pseudo
[d6/06dd54] Submitted process > run_augustus_contigs
[62/d6e281] Submitted process > run_augustus_pseudo
[ce/16047c] Submitted process > merge_genemodels
[bc/15066e] Submitted process > integrate_genemodels
[2a/62c852] Submitted process > remove_exons
[29/5de8c9] Submitted process > pseudogene_calling (1)
[59/299643] Submitted process > merge_structural (1)
[a0/91e559] Submitted process > add_gap_features (1)
[76/596d97] Submitted process > split_splice_models_at_gaps (1)
[d1/9fc69a] Submitted process > add_polypeptides (1)
[e3/e512b3] Submitted process > get_proteins_for_orthomcl (1)
[b4/bbe10b] Submitted process > run_pfam (1)
[5b/2db645] Submitted process > run_pfam (2)
[a0/fc6465] Submitted process > run_pfam (3)
[47/d2bcac] Submitted process > run_pfam (4)
[80/91a1eb] Submitted process > run_pfam (5)
[ee/718eb8] Submitted process > run_pfam (7)
[e5/dfd191] Submitted process > run_pfam (9)
[10/d2ce74] Submitted process > run_pfam (10)
[d4/660d23] Submitted process > run_pfam (6)
[8e/6dd46c] Submitted process > run_pfam (8)
[60/c9eabb] Submitted process > make_target_input_for_orthomcl (1)
[32/fa2625] Submitted process > run_pfam (11)
[b8/ab63ae] Submitted process > run_pfam (12)
[49/4414d6] Submitted process > run_pfam (13)
[bf/533f9a] Submitted process > run_pfam (14)
[4c/a9c07a] Submitted process > run_pfam (15)
[e8/22b26b] Submitted process > run_pfam (16)
[82/72e91c] Submitted process > run_pfam (17)
[71/da9ef1] Submitted process > run_pfam (18)
[21/0b9bb8] Submitted process > run_pfam (19)
[00/7fe92b] Submitted process > run_pfam (20)
[6b/ebf599] Submitted process > run_pfam (21)
[6c/cc06c6] Submitted process > run_pfam (22)
[58/70ec64] Submitted process > run_pfam (23)
[19/149b3c] Submitted process > run_pfam (24)
[fd/2deb4a] Submitted process > run_pfam (25)
[ff/8fb6b5] Submitted process > run_pfam (26)
[77/b7ad18] Submitted process > run_pfam (27)
[a7/33127b] Submitted process > run_pfam (28)
[fd/688841] Submitted process > run_pfam (29)
[6d/e06fc0] Submitted process > run_pfam (30)
[51/f5d058] Submitted process > run_pfam (31)
[ba/55e216] Submitted process > run_pfam (32)
[e2/d97868] Submitted process > run_pfam (33)
[d8/809174] Submitted process > run_pfam (34)
[c3/229b84] Submitted process > run_pfam (35)
[e3/c04bbd] Submitted process > run_pfam (36)
[61/e902c5] Submitted process > run_pfam (37)
[a5/dbcf87] Submitted process > run_pfam (38)
[9e/35761e] Submitted process > run_pfam (39)
[2f/6fffde] Submitted process > run_pfam (40)
[37/bb8fe5] Submitted process > run_pfam (41)
[08/db849f] Submitted process > run_pfam (42)
[86/ccb927] Submitted process > run_pfam (43)
[01/4267e5] Submitted process > run_pfam (44)
[76/6f75fe] Submitted process > run_pfam (45)
[d5/a9e743] Submitted process > run_pfam (46)
[45/c028c4] Submitted process > run_pfam (47)
[ae/b42530] Submitted process > run_pfam (48)
[67/eda7e7] Submitted process > run_pfam (49)
[f5/877ab8] Submitted process > run_pfam (50)
[d2/ef8a32] Submitted process > run_pfam (51)
[d9/5600db] Submitted process > run_pfam (52)
[b3/8e4ba9] Submitted process > run_pfam (53)
[c7/3bbe1e] Submitted process > run_pfam (54)
[f8/40596e] Submitted process > run_pfam (55)
[fb/01755d] Submitted process > run_pfam (56)
[b3/227472] Submitted process > run_pfam (57)
[95/987df9] Submitted process > run_pfam (58)
[44/90c588] Submitted process > run_pfam (59)
[97/8e3bc2] Submitted process > run_pfam (60)
[02/d392f1] Submitted process > run_pfam (61)
[24/e5bbc9] Submitted process > run_pfam (62)
[ea/e9634a] Submitted process > run_pfam (63)
[44/4c3828] Submitted process > run_pfam (64)
[9b/d004fb] Submitted process > run_pfam (65)
[71/854734] Submitted process > run_pfam (66)
[3e/722ca7] Submitted process > run_pfam (67)
[51/616316] Submitted process > run_pfam (68)
[0f/345b2e] Submitted process > run_pfam (69)
[1a/283104] Submitted process > run_pfam (70)
[c5/3c63bb] Submitted process > run_pfam (71)
[81/a749ec] Submitted process > run_pfam (72)
[f1/bccf12] Submitted process > run_pfam (73)
[56/028008] Submitted process > run_pfam (74)
[45/516a0f] Submitted process > run_pfam (75)
[43/9a4796] Submitted process > run_pfam (76)
[2d/3b2e23] Submitted process > run_pfam (77)
[c7/1c724f] Submitted process > run_pfam (78)
[55/99be77] Submitted process > run_pfam (79)
[39/ff6e3f] Submitted process > run_pfam (80)
[d8/2601b4] Submitted process > run_pfam (81)
[94/180e6e] Submitted process > run_pfam (82)
[bc/2e97a6] Submitted process > run_pfam (83)
[1c/3047b4] Submitted process > run_pfam (84)
[f8/cb5934] Submitted process > run_pfam (85)
[0b/e7aaf3] Submitted process > run_pfam (86)
[ac/2231fc] Submitted process > run_pfam (87)
[cb/bccafd] Submitted process > run_pfam (88)
[d3/3d10bd] Submitted process > run_pfam (89)
[95/0d60b4] Submitted process > run_pfam (90)
[f4/0e492a] Submitted process > run_pfam (91)
[af/e91fcb] Submitted process > run_pfam (92)
[e0/230784] Submitted process > run_pfam (93)
[c4/00969a] Submitted process > run_pfam (94)
[00/5ae98f] Submitted process > run_pfam (95)
[e5/721f3a] Submitted process > run_pfam (96)
[35/f8dcd8] Submitted process > run_pfam (97)
[ac/37811d] Submitted process > run_pfam (98)
[83/c6d4cd] Submitted process > run_pfam (99)
[29/6b9877] Submitted process > run_pfam (100)
[27/72f700] Submitted process > run_pfam (101)
[7c/133b9b] Submitted process > run_pfam (102)
[69/2c97be] Submitted process > run_pfam (103)
[7b/305f47] Submitted process > run_pfam (104)
[96/5f1658] Submitted process > run_pfam (105)
[bd/7aab7a] Submitted process > run_pfam (106)
[c1/d11a9f] Submitted process > run_pfam (107)
[21/4c9d0f] Submitted process > run_pfam (108)
[99/137ff5] Submitted process > run_pfam (109)
[b2/336c3d] Submitted process > run_pfam (110)
[ce/4aade0] Submitted process > run_pfam (111)
[b1/a33b9f] Submitted process > run_pfam (112)
[7c/504798] Submitted process > run_pfam (113)
[15/babcbe] Submitted process > run_pfam (114)
[e6/257e90] Submitted process > run_pfam (115)
[2e/000930] Submitted process > run_pfam (116)
[fa/88b968] Submitted process > run_pfam (117)
[62/b7d019] Submitted process > run_pfam (118)
[da/723c6f] Submitted process > run_pfam (119)
[09/40c576] Submitted process > run_pfam (120)
[8e/7df6cb] Submitted process > run_pfam (121)
[6b/0d2f5d] Submitted process > run_pfam (122)
[51/ddbedc] Submitted process > run_pfam (123)
[0f/87370e] Submitted process > run_pfam (124)
[47/fd6a43] Submitted process > run_pfam (125)
[ea/1ebf8e] Submitted process > run_pfam (126)
[4e/c71fd7] Submitted process > run_pfam (127)
[d7/f5a4f1] Submitted process > run_pfam (128)
[2e/9eb534] Submitted process > run_pfam (129)
[94/0b0d8a] Submitted process > run_pfam (130)
[82/67a541] Submitted process > run_pfam (131)
[f2/72c4e2] Submitted process > run_pfam (132)
[c7/1ded54] Submitted process > run_pfam (133)
[3d/c4cbee] Submitted process > run_pfam (134)
[af/23116b] Submitted process > run_pfam (135)
[ef/6922e9] Submitted process > run_pfam (136)
[0d/67af6f] Submitted process > run_pfam (137)
[c3/130608] Submitted process > run_pfam (138)
[64/5339d3] Submitted process > run_pfam (139)
[fe/f6a0e1] Submitted process > run_pfam (140)
[a2/1bb7d0] Submitted process > run_pfam (141)
[30/3a6219] Submitted process > run_pfam (142)
[70/fe21d9] Submitted process > run_pfam (143)
[21/da0283] Submitted process > run_pfam (144)
[f0/6408a2] Submitted process > run_pfam (145)
[9a/eed91e] Submitted process > run_pfam (146)
[b9/061041] Submitted process > run_pfam (147)
[a3/e60624] Submitted process > run_pfam (148)
[44/c19cb4] Submitted process > run_pfam (149)
[2c/ec5768] Submitted process > run_pfam (150)
[0f/d32546] Submitted process > run_pfam (151)
[7a/c7f339] Submitted process > run_pfam (152)
[1c/189b11] Submitted process > run_pfam (153)
[90/31c39f] Submitted process > run_pfam (154)
[20/8ae8fa] Submitted process > run_pfam (155)
[81/ff5004] Submitted process > run_pfam (156)
[92/af9246] Submitted process > run_pfam (157)
[32/65325d] Submitted process > run_pfam (158)
[a7/d3f16c] Submitted process > run_pfam (159)
[a2/894c68] Submitted process > run_pfam (160)
[51/275860] Submitted process > run_pfam (161)
[03/dbb10a] Submitted process > run_pfam (162)
[73/435c91] Submitted process > run_pfam (163)
[37/1aec38] Submitted process > run_pfam (164)
[a2/d3d757] Submitted process > run_pfam (165)
[98/79a42f] Submitted process > run_pfam (166)
[d2/8e3d04] Submitted process > run_pfam (167)
[5e/d3827f] Submitted process > run_pfam (168)
[14/40dae4] Submitted process > run_pfam (169)
[df/e18164] Submitted process > run_pfam (170)
[a8/e906ef] Submitted process > run_pfam (171)
[38/f88edf] Submitted process > run_pfam (172)
[01/1ece7c] Submitted process > run_pfam (173)
[4f/ffac37] Submitted process > run_pfam (174)
[5e/8e88a5] Submitted process > run_pfam (175)
[16/75add5] Submitted process > run_pfam (176)
[f7/55d712] Submitted process > run_pfam (177)
[02/f06e91] Submitted process > run_pfam (178)
[b5/b88cc6] Submitted process > run_pfam (179)
[6b/da77d7] Submitted process > run_pfam (180)
[31/ab620e] Submitted process > run_pfam (181)
[aa/9fe143] Submitted process > run_pfam (182)
[a0/4b7608] Submitted process > run_pfam (183)
[12/f0edc1] Submitted process > run_pfam (184)
[0c/57879a] Submitted process > run_pfam (185)
[29/2eae91] Submitted process > run_pfam (186)
[83/09ac7c] Submitted process > run_pfam (187)
[a9/6e4f4a] Submitted process > run_pfam (188)
[fd/57f0b3] Submitted process > run_pfam (189)
[60/140cf8] Submitted process > run_pfam (190)
[78/e7c6c6] Submitted process > run_pfam (191)
[21/814953] Submitted process > run_pfam (192)
[58/7847ae] Submitted process > run_pfam (193)
[75/dac202] Submitted process > run_pfam (194)
[0b/8c9ae4] Submitted process > run_pfam (195)
[86/83eb37] Submitted process > run_pfam (196)
[47/0c2d33] Submitted process > run_pfam (197)
[84/ef2184] Submitted process > run_pfam (198)
[36/5c3e5b] Submitted process > run_pfam (199)
[ec/6a644b] Submitted process > run_pfam (200)
[56/4e6602] Submitted process > run_pfam (201)
[78/f8fec6] Submitted process > run_pfam (202)
[46/13232e] Submitted process > run_orthofinder (1)
[ee/193264] Submitted process > pfam_to_gff3 (1)
[b2/5dfd3b] Submitted process > annotate_orthologs (1)
[b9/046ba7] Submitted process > annotate_pfam (1)
[e9/c225ea] Submitted process > make_distribution_gff (1)
[c8/866f90] Submitted process > make_distribution_gaf (1)
[82/6b4ed1] Submitted process > make_genome_stats (1)
[50/aed626] Submitted process > add_products_to_protein_fasta (1)
[1b/38f203] Submitted process > merge_gff3_for_gff3toembl (1)
[f2/ec88f8] Submitted process > make_genelist (1)
[7a/b5af20] Submitted process > make_report (1)
[a4/0d3fbb] Submitted process > reference_compare (1)
[7e/2aa50b] Submitted process > make_circos_inputs (1)
[6f/057f7d] Submitted process > make_embl (1)
[5f/ec4f61] Submitted process > make_tree (1)
[7c/75d10e] Submitted process > circos_run_chrs (5A)
[42/9c2024] Submitted process > circos_run_chrs (7A)
[84/c00db7] Submitted process > circos_run_chrs (RA)
[d3/974e47] Submitted process > circos_run_chrs (6A)
[99/354357] Submitted process > circos_run_chrs (3A)
[2e/963617] Submitted process > circos_run_chrs (4A)
[2e/deb94f] Submitted process > circos_run_chrs (2A)
[01/252590] Submitted process > circos_run_chrs (1A)
| Pseudochromosomes | ✓ |
| Protein evidence | ✗ |
| BRAKER | ✓ |
| Transfer tool | RATT |
| RATT transfer type | Species |
| SNAP | ✗ |
| Max gene length | 50000 |
| Max overlap | |
| Score threshold | 0.4 |
| Transcript evidence | ✗ |
Information available in the result tabs
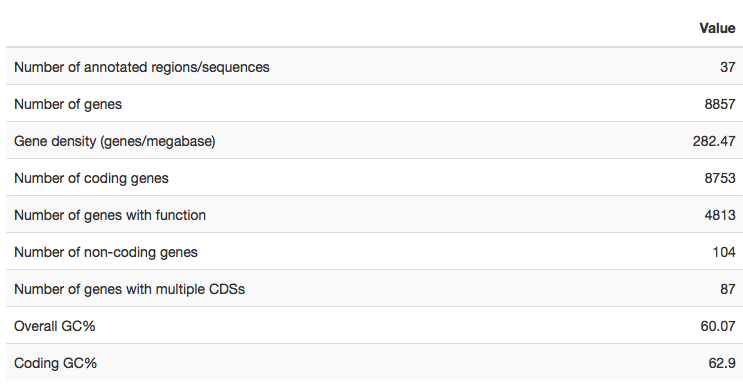
Genome statistics
The Genome statistics tab summarizes basic numeric statistics about the annotated genome, such as
- gene counts
- gene density
- GC content
This summary is useful to quickly assess the success of the annotation job: does the amount of annotated features match the expectations?
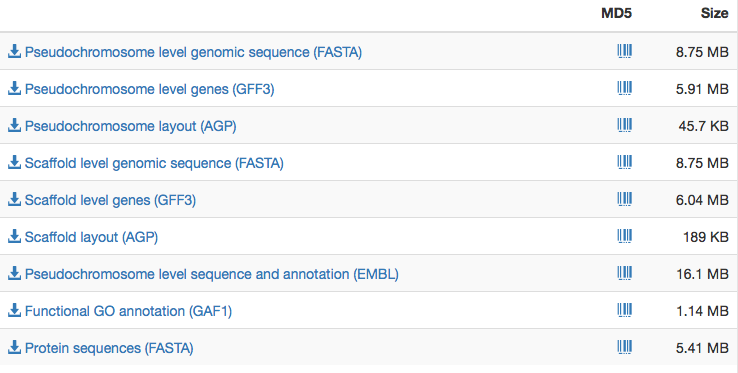
Result files
The next tab (Result files) allows the user to download the results of the annotation jobs to their own computer, e.g. for further downstream analyses, manual curation or preparation for database submission. These are offered in the following formats:
- GFF3 and EMBL format for annotated features
- FASTA format for sequences
- GAF format for GO terms for each functionally annotated gene
- AGP format files for pseudochromosome layouts
If no pseudochromosome contiguation was selected, files for both levels are identical. We also calculate MD5 tags which may be required at the time of submission in case there is no need for further curation.
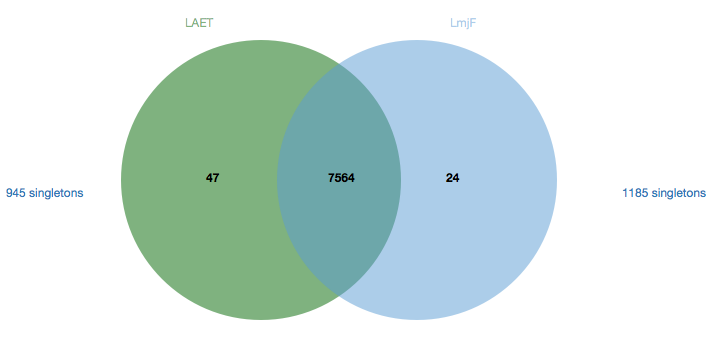
Orthology
The Venn diagram on the Orthology tab depicts shared gene cluster membership between the newly annotated genome and the reference to easily assess the amount of species-specific gene content. The set on the left (green) represents the target genome while the one on the right (blue) represents the reference. The numbers in the set circles specify the number of shared or species-specific clusters with at least two genes. The amount of genes not appearing in any cluster (singletons) is shown outside both sets.
By clicking on the numbers in the diagram, it is possible to browse the content of all sets using interactive tables. These paginated and searchable tables are shown below the Venn diagram when a number is clicked. A flat text file (in OrthoMCL 2.0 format) is also provided for custom analyses.
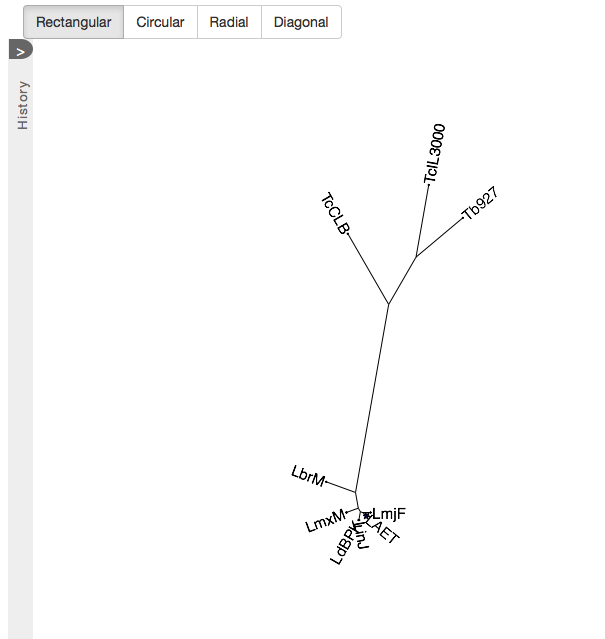
Phylogeny
The tree on the Phylogeny tab shows how the newly annotated target genome fits into the phylogeny of related species. From a set of up to 50 single-copy genes shared among all these species, a quick tree is built and visualized in an interactive tree, supporting various tree styles. All data used to build the tree are also available for download.
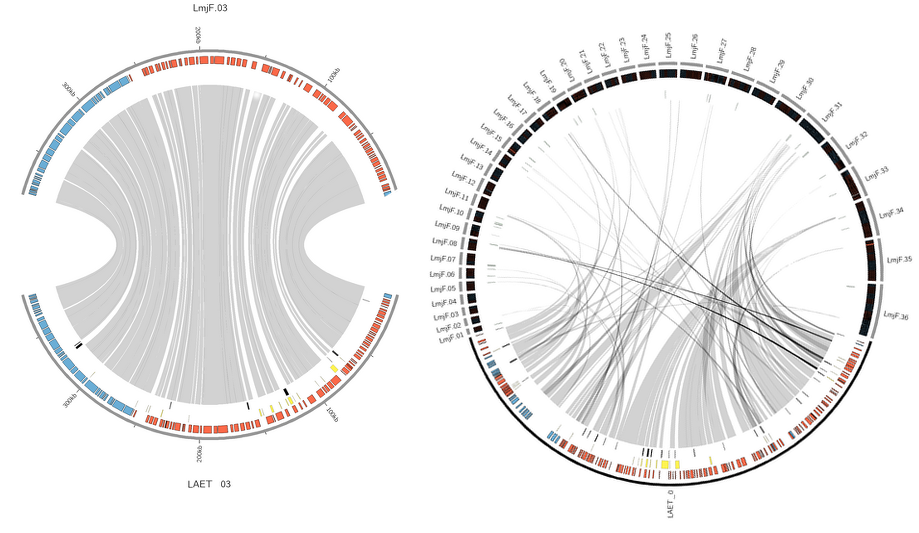
Synteny
The Synteny tab contains circular plots showing alignments between each reference chromosome and their newly annotated counterpart. The reference chromosome is shown at the top and the new pseudochromosome is shown at the bottom. Grey ribbons between both represent similar regions as identified by BLASTN matches. Genes on the forward strand (blue) and the reverse strand (red) are annotated on the chromosomes, as well as gaps (yellow), singletons (black) and missing core genes (green), each in separate tracks. A similar output is created for uncontiguated input sequences concatenated into a 'bin' sequence (not drawn to scale) compared to all reference chromosomes.
These plots are useful to visualize the reference-target colinearity, which helps in identifying potential problems in pseudochromosome contiguation or large-scale chromosomal rearrangements at a glance.
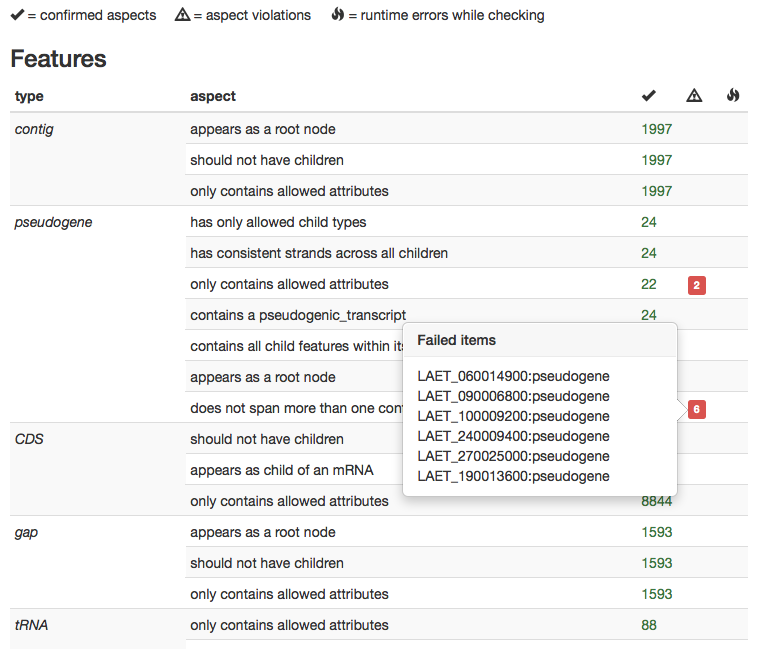
Validator results
We are doing our best to ensure that gene annotation files delivered by Companion are both syntactically and semantically (=biologically) valid and do not contain major issues. However, due to the complexity of the process and the high variability of input data, it is sometimes possible to end up with less than perfect gene models. Often such remaining issues can be quickly addressed by a human curator.
To make the best use of curator time as a scarce resource, we perform thorough automatic checking of our generated annotations, given as annotation graphs in GFF3 format. The results of this check are presented in a table, grouped by feature type. The number of successful and failed checks is concisely presented and for each failed check, the ID of the questionable feature is reported by clicking on the red labels denoting checking failures. This makes it easy to quickly estimate the amount of required curation and allows to work in a directed and focused fashion.







